Oct 26, 2017
Using pip on a Debian/Ubuntu system can result in this error: TypeError:
unsupported operand type(s) for -=: 'Retry' and 'int'. E.g. see this error
report in Debian: https://bugs.debian.org/cgi-bin/bugreport.cgi?bug=870701
The problem seems to be that requests/pip vendors some libraries and
Debian/Ubuntu un-vendors them again. This results in the same code imported
multiple times under different names. Funnily the above exception results from
such a mismatch if a network request has an error/ times out. I tried to
follow the explanaition why this results in the above
error, but gave up. :-)
Anyway: we use python3.6 -m venv ... to create virtualenvs and afterwards
pip to install requirements into the virtualenv. Sometimes the error happened
and I couldn’t finish the installations even after multiple tries. As the
problem was that pip uses an un-vendored version of the requests library, the
easiest way to fix it was to switch pip back to use a vendored requests
library:
# this assumes the virtualenv is created in '.venv'
# This will copy the unbundled versions of the libs from /usr/share/python-wheels
# into .venv/share/python-wheels
(md .venv && cd .venv && $(SYSTEM_PYTHON36) -m venv --copies .)
# install a clean copy of requests...
# will also install chardet/idna/urllib3/certifi and so on into .venv
# this might stil fail with the above error...
.venv/bin/pip install requests wheel
# remove the unbundled versions of the libs
(cd .venv/share/python-wheels/ && rm requests-*.whl chardet-*.whl urllib3-*.whl)
Afterwards pip install --requirement=requirements.txt now succeeded.
If you also need a system pip, follow this instructions:
https://stackoverflow.com/a/37531821/1380673)
Oct 23, 2016
Since a few years, pipelines (via %>% of the magrittr package) are quite popular in R and the grown ecosystem of the “tidyverse” is built around pipelines. Having tried both the pandas syntax (e.g. chaining like df.groupby().mean() or plain function2(function1(input))) and the R’s pipeline syntax, I have to admit that I like the pipeline syntax a lot more.
In my opinion the strengths of R’s pipeline syntax are:
- The same verbs can be used for different inputs (there are SQL backends for dplyr), thanks to R’s single-dispatch mechanism (called S3 objects).
- Thanks to using function instead of class methods, it’s also more easily extendable (for a new method on
pandas.DataFrame you have to add that to the pandas repository or you need to use monkey patching). Fortunatelly, both functions and singledispatch are also available in python :-)
- It uses normal functions as pipline parts:
input %>% function() is equivalent to function(input). Unfortunately, this isn’t easily matched in python, as pythons evaluation rules would first evaluate function() (e.g. call functions without any input). So one has to make function() return a helper object which can then be used as a pipeline part.
- R’s delayed evaluation rules make it easy to evaluate arguments in the context of the pipeline, e.g.
df %>% select(x) would be converted to the equivalent of pandas df[["x"]], e.g. the name of the variable will be used in the selection. In python it would either error (if x is not defined) or (if x was defined, e.g. x = "column"), would take the value of x, e.g. df[["column"]]. For this, some workarounds exist by using helper objects like select(X.x), e.g. pandas-ply and its Symbolic expression.
There exist a few implementation of dplyr like pipeline verbs for python (e.g. pandas itself, pandas-ply (uses method chaining instead of a pipe operator), dplython, and dfply), but they all focus on implementing dplyr style pipelines for pandas.DataFrames and I wanted to try out a simpler but more general approach to pipelines.
The code
The following shows my take on how to implement the first three things (I left out “Symbolic expressions”). The code is available in https://github.com/janschulz/pydatapipes. The short (removed the docstrings) version is actually only a few lines of code:
from functools import singledispatch, wraps
class PipeVerb():
"""Object which represents a part of a pipeline"""
def __init__(self, func, *args, **kwargs):
self.pipe_func = func
self.args = args
self.kwargs = kwargs
def __rrshift__(self, input):
return self.pipe_func(input, *self.args, **self.kwargs)
def pipeverb(func):
"""Decorator to convert a function to a pipeline verb (without singledispatch)"""
@wraps(func)
def decorated(*args, **kwargs):
return PipeVerb(func, *args, **kwargs)
# If it is a singledispatch method, expose the register method here as well
if hasattr(func, 'register'):
decorated.register = func.register
return decorated
def make_pipesource(cls):
"""Enables a class to function as a pipe source"""
if hasattr(cls, '__rshift__') and (not getattr(cls.__rshift__, 'pipeoperator', False)):
def __rshift__(self, other):
"""Pipeline operator if the right side is a PipeVerb"""
if isinstance(other, PipeVerb):
return other.__rrshift__(self)
else:
return self.__orig_rshift__(other)
cls.__orig_rshift__ = cls.__rshift__
cls.__rshift__ = __rshift__
setattr(cls.__rshift__, "pipeoperator", True)
def singledispatch_pipeverb(func):
"""Convenience decorator to convert a function to a singledispatch pipeline verb"""
return pipeverb(singledispatch(func))
Simple pipeline verbs
For end users wanting to build a new pipeline verb or add pipeline functionality to a new data source,
there are two functions to build new pipeline parts:
#from pydatapipes.pipes import singledispatch_pipeverb, make_pipesource
import pandas as pd
# generic version which defines the API and should raise NotImplementedError
@singledispatch_pipeverb
def append_col(input, x = 1):
"""Appends x to the data source"""
raise NotImplementedError("append_col is not implemented for data of type %s" % type(input))
# concrete implementation for pandas.DataFrame
@append_col.register(pd.DataFrame)
def append_col_df(input, x = 1):
# always ensure that you return new data!
copy = input.copy()
copy["X"] = x
return copy
# ensure that pd.DataFrame is usable as a pipe source
make_pipesource(pd.DataFrame)
This can then be used in a pipeline:
import pandas as pd
print(pd.DataFrame({"a" : [1,2,3]}) >> append_col(x=3))
The above example implements a pipeline verb for pandas.DataFrame, but due to the useage of
singledispatch, this is generic. By implementing additional
append_col_<data_source_type>() functions and registering it with the original append_col function,
the append_col function can be used with other data sources, e.g. SQL databases, HDF5, or even builtin data
types like list or dict:
@append_col.register(list)
def append_col_df(input, x = 1):
return input + [x]
[1, 2] >> append_col()
If a verb has no actual implementation for a data source, it will simply raise an NotImplementedError:
try:
1 >> append_col()
except NotImplementedError as e:
print(e)
append_col is not implemented for data of type <class 'int'>
A more complex example: grouped and ungrouped aggregation on DataFrames
singledispatch also makes it easy to work with grouped and ungrouped pd.DataFrames:
@singledispatch_pipeverb
def groupby(input, columns):
"""Group the input by columns"""
raise NotImplementedError("groupby is not implemented for data of type %s" % type(input))
@groupby.register(pd.DataFrame)
def groupby_DataFrame(input, columns):
"""Group a DataFrame"""
return input.groupby(columns)
@singledispatch_pipeverb
def summarize_mean(input):
"""Summarize the input via mean aggregation"""
raise NotImplementedError("summarize_mean is not implemented for data of type %s" % type(input))
@summarize_mean.register(pd.DataFrame)
def summarize_mean_DataFrame(input):
"""Summarize a DataFrame via mean aggregation"""
return input.mean()
@summarize_mean.register(pd.core.groupby.GroupBy)
def summarize_mean_GroupBy(input):
"""Summarize a grouped DataFrame via mean aggregation"""
return input.mean()
df = pd.DataFrame({"a" : [1, 2, 3, 4], "b": [1, 1, 2, 2]})
print(df >> summarize_mean())
a 2.5
b 1.5
dtype: float64
print(df >> groupby("b") >> summarize_mean())
Limitiations
Compared to R’s implementation in the magrittr package,
input >> verb(x) can’t be rewritten as verb(input, x).
The problem here is that verb(x) under the hood constructs a helper object (PipeVerb) which
is used in the rshift operation. At the time of calling verb(...), we can’t always be sure
whether we want an object which can be used in the pipeline or want to already
compute the result. As an example consider a verb merge(*additional_data). You could call
that as data >> merge(first, second) to indicate that you want all three (data,
first, and second) merged. On the other hand, merge(first, second) is also valid
(“merge first and second together).
Usage as function and pipeline verb
To help work around this problem, the convenience decorator singledispatch_pipeverb is actually not the best option if
you want to create reusable pipeline verbs. Instead, the singledispatch_pipeverb decorator is also available in
two parts, so that one can both expose the original function (with singledispatch enabled) and the
final pipeline verb version:
#from pydatapipes.pipes import pipeverb, singledispatch
# first use singledispatch on the original function, but define it with a trailing underscore
@singledispatch
def my_verb_(input, x=1, y=2):
raise NotImplemented("my_verb is not implemented for data of type %s" % type(input))
# afterwards convert the original function to the pipeline verb:
my_verb = pipeverb(my_verb_)
# concrete implementations can be registered on both ``my_verb`` and ``my_verb_``
@my_verb_.register(list)
def my_verb_df(input, x=1, y=2):
return input + [x, y]
A user can now use both versions:
Rules and conventions
To work as a pipline verb, functions must follow these rules:
- Pipelines assume that the verbs itself are side-effect free, i.e. they do not change the inputs of
the data pipeline. This means that actual implementations of a verb for a specific data source
must ensure that the input is not changed in any way, e.g. if you want to pass on a changed value
of a
pd.DataFrame, make a copy first.
- The initial function (not the actual implementations for a specific data source) should usually
do nothing but simply raise
NotImplementedError, as it is called for all other types of data
sources.
The strength of the tidyverse is it’s coherent API design. To ensure a coherent API for pipeline verbs,
it would be nice if verbs would follow these conventions:
- Pipeline verbs should actually be named as verbs, e.g. use
input >> summarize() instead of
input >> Summary()
- If you expose both the pipeline verb and a normal function (which can be called directly),
the pipeline verb should get the “normal” verb name and the function version should get
an underscore
_ appended: x >> verb() -> verb_(x)
- The actual implementation function of a
verb() for a data source of class Type
should be called verb_Type(...), e.g. select_DataFrame()
Missing parts
So what is missing? Quite a lot :-)
- Symbolic expressions: e.g.
select(X.x) instead of select("x")
- Helper for dplyr style column selection (e.g.
select(starts_with("y2016_")) and select(X[X.first_column:X.last_column]))
- all the dplyr, tidyr, … verbs which make the tidyverse so great
Some of this is already implemented in the other dplyr like python libs (pandas-ply, dplython, and dfply), so I’m not sure how to go on. I really like my versions of pipelines but duplicating the works of them feels like a waste of time. So my next step is seeing if it’s possible to integrate this with one of these solutions, probably dfply as that looks the closest implementation.
[This post is also available as a jupyter notebook]
Jan 26, 2016
Recently someone was surprised that I use windows as my main dev machine as other OS usually are developer friendly. Out of the box, this is true. But to make yourself at home as a developer, you usually change a lot of things, no matter if you are using OS X, Linux or Win. So here is what I use:
- proper command line: cmder with git
- Pycharm + Notepad++ as editor
- python from miniconda with multiple envs
- jupyter notebook with a conda env kernel manager
Not all is windows specific… I actually suspect that a lot is windows agnostic and I would use a similar setup on a different OS…
A proper command line: cmder
Windows cmd is pretty limited, both because there is almost no commands available and because of the terminal window itself lacks tab competition, history, proper C&P… I use cmder as a replacement. Use the upcoming 1.3 version, it changes the way the config / startup files are handled -> available as an artifact in the Appveyor builds (e.g. this one). It comes with better tab completion (including for git commands), history, search previous commands, c&p, git integration in the prompt, and can be customized via a startup profile. It also includes a copy of git for windows 2.x, so for most case, there is no need to install git by yourself. You can use cmd, bash (comes with the copy of git) and powershell.
I install it in a dropbox subfolder, which means that I have the same environment even at work. Run cmder.exe /REGISTER ALL once as admin to get the cmder here item in the right click menu in windows explorer.
In config\user-profile.cmd, I add a few more path items and also start an ssh agent:
:: needs the private ssh key in %USERPROFILE%\.ssh\
@call start-ssh-agent
:: add my own scripts
@set "PATH=%PATH%;%CMDER_ROOT%\vendor\jasc"
:: add unix commands from existing git -> last to not shadow windows commands...
@set "PATH=%PATH%;%GIT_INSTALL_ROOT%\usr\bin\"
Thanks to the last line, I’ve ls, grep, find, ssh, … available in the command line.
Aliases are in config\aliases. I add things like
w=where $1
cdp=cd c:\data\external\projects
ls_envs=ls c:\portabel\miniconda\envs\
note="C:\Program Files (x86)\Notepad++\notepad++.exe" $*
I also customize the prompt (via a config/conda.lua file) so that activating a conda env will show up in the prompt (The need for the reset is IMO a bug):
---
-- Find out the basename of a file/directory (last element after \ or /
-- @return {basename}
---
function basename(inputstr)
sep = "\\/"
local last = nil
local t={} ; i=1
for str in string.gmatch(inputstr, "([^"..sep.."]+)") do
--t[i] = str
--i = i + 1
last = str
end
return last
end
---
-- Find out if the String starts with Start
-- @return {boolean}
---
function string.starts(String,Start)
return string.sub(String,1,string.len(Start))==Start
end
---
-- Find out current conda env
-- @return {false|conda env name}
---
function get_conda_env()
env_path = clink.get_env('CONDA_DEFAULT_ENV')
if env_path then
basen = basename(env_path)
return basen
end
return false
end
---
-- after conda activate: reset prompt
---
function reset_prompt_filter()
-- reset to original, e.g. after conda activate destroyed it...
if string.match(clink.prompt.value, "{lamb}") == nil or not string.starts(clink.prompt.value,"\x1b[") then
-- orig: $E[1;32;40m$P$S{git}{hg}$S$_$E[1;30;40m{lamb}$S$E[0m
-- color codes: "\x1b[1;37;40m"
cwd = clink.get_cwd()
prompt = "\x1b[1;32;40m{cwd} {git}{hg} \n\x1b[1;30;40m{lamb} \x1b[0m"
new_value = string.gsub(prompt, "{cwd}", cwd)
clink.prompt.value = new_value
end
end
---
-- add conda env name
---
function conda_prompt_filter()
-- add in conda env name
local conda_env = get_conda_env()
if conda_env then
clink.prompt.value = string.gsub(clink.prompt.value, "{lamb}", "["..conda_env.."] {lamb}")
end
end
clink.prompt.register_filter(reset_prompt_filter, 10)
clink.prompt.register_filter(conda_prompt_filter, 20)
local function tilde_match (text, f, l)
if text == '~' then
clink.add_match(clink.get_env('userprofile'))
clink.matches_are_files()
return true
end
end
clink.register_match_generator(tilde_match, 1)
git setup
I usually add two remotes: the upstream repo as origin (using the https URL for git clone) and my fork as mine (using the ssh URL for git remote add mine <ssh-url>). I do that even in cases where I am the upstream.
mine is setup as the default remote push location and git push defaults to the current branch. That way I can do things like git push without specifying a remote or without getting a confirmation message on first push of a branch.
Thanks to the ssh agent started by cmder on startup, I only have to give my password once per session.
I’ve setup notepad as the git commit editor but probably will switch to Sublime Text because of the better spell checking…
The following are the relevant lines of my %USERPROFILE%\.gitconfig:
[...]
[core]
editor = \"C:\\Program Files (x86)\\Notepad++\\notepad++.exe\" -multiInst -nosession -noPlugin
excludesfile = ~/.gitignore-global # for things like the .idea dir from pycharm
[push]
# don't show a setup message on first push of the branch
default = current
[remote]
# per default push to "mine"
pushdefault = mine
[alias]
unadd = reset HEAD --
fixup = commit --amend --no-edit
slog = log --pretty=oneline --abbrev-commit
dc = diff --cached
# specially for word files which are shown as text in the latest git for windows 2.x builds
wd = diff --word-diff
I also install git-extras, mainly for git pr (checkout a github PR directly from origin), git ignore, git changelog
Python development: editors, conda
Editors: Pycharm, Notepad++, Sublime Text 3
I mainly use a combination of Pycharm (IDE for bigger projects/changes), Notepad++ (small patches, build related stuff) and recently Sublime Text 3 (replacement for notepad++, lets see…). Notepad++ is setup to replace notepad.exe, so anything which calls notepad will bring up Notepad++. Other than that, I use no special config for the IDE/editors…
conda python
I currently use a miniconda py27 setup (which I should update to a py3.x based one, but am too lazy…), but use envs for most of the work (e.g. the main env has mostly only conda + conda build related stuff in it). The default env is added to the default path (either by the installer or by using setx path C:\portabel\miniconda;C:\portabel\miniconda\Scripts;%PATH% in a cmd, not cmder window). I create additional envs with conda create -n <env-name> python=x.x pandas matplotlib ... as needed. Pycharm can use envs as additional interpreters, so no problem there… On the command line, thanks to the above cmder setup, an ls_envs will show all environments and activate <env-name> works without problems and the conda env name is shown in the command line.
I installed the visual studio compilers for 2.7, 3.4 and 3.5 by religiously following the following blog post on “Compiling Python extensions on Windows” by @ionelmc. It works!
If conda has no package for the package you want, activate the env, conda install pip and then use pip to install the package into that env. conda list shows both conda packages and pip packages.
Jupyter notebook
I have one “jupyter-notebook” env which holds the install for the notebook (e.g. conda create -n jupyter-notebook python=3.5 notebook). I start notebook servers via shortcuts, which point to the jupyter-notebook.exe entry in the jupyter-notebook env (e.g. C:\portabel\miniconda\envs\jupyter-notebook\Scripts\jupyter-notebook.exe) and which are setup to start in the main project directory (e.g. c:\data\external\projects\projectA\). That way I can startup multiple notebook servers in different project dirs by using multiple shortcuts.
Add all conda envs as kernels
I use Cadair/jupyter_environment_kernels (with an additional PR) as a kernel manager, so all my conda environments show up as additional kernel entries. For each project, I setup a new conda environment which is then used in the project notebooks as kernel.
Add-ons for jupyter notebook
I install the jupyter notebook extensions (installed in the jupyter-notebook conda environment), mainly for the Table of Content support.
I also add some ipython magic commands to each python environment which is used as notebook kernel:
proper diffs and commits for notebooks
I usually don’t want to commit the outputs of a notebook to git, so I strip them with a git clean filter.
I also want git diff to show something which I can actually read instead of the raw json file content, so I also setup a special converter which is used by git diff before comparing the files.
There are a lot of scripts around for that, but most use python (e.g. strip output (gist, kynan/nbstripout) and nbflatten but this is slow for big notebooks. :-( Fortunately, the nbflatten gist also introduced me to jq, something like grep and sed/awk for json data. After sorting out a windows bug, this jq based nbflatten script now works on windows, too. Below is a slightly adjusted variant of that script.
This needs a recent jq.exe (>=1.6, not yet released, go to https://ci.appveyor.com/project/stedolan/jq and click on one of the passing builds -> 64bit -> ARTIFACTS) due to a bug in 1.5. Put jq.exe in your path (e.g. <cmder>\bin) and add the following file somewhere:
# based on https://gist.github.com/jfeist/cd00aa3b681092e1d5dc
def banner: "\(.) " + (28-(.|length))*"-";
# metadata
("Non-cell info" | banner), del(.cells), "",
# content
(.cells[] | (
("\(.cell_type) cell" | banner),
(.source[] | rtrimstr("\n")), # output source
if ($show_output == "1") then # the cell output only when it is requested..
"",
(select(.cell_type=="code" and (.outputs|length)>0) | (
("output" | banner),
(.outputs[] | (
(select(.text) | "\(.text|add)" | rtrimstr("\n")),
(select(.traceback) | (.traceback|join("\n"))),
(select(.text or .traceback|not) | "(Non-plaintext output)")
)
),
""
)
)
else
""
end
)
)
I put mine as nbflatten.jq into a cmder subdir.
I now have the following in my global .gitconfig:
[filter "stripoutput"]
# removes output and execution counts form the notebook before committing it
clean = "jq --indent 1 '(.cells[] | select(has(\"outputs\")) | .outputs) = [] | (.cells[] | select(has(\"execution_count\")) | .execution_count) = null'"
[diff "ipynb"]
# uses a "flattend" representation of the notebook for diffing
# note the quotes and the escapes for the quotes around the filename and the backslashes...
textconv = "jq -r -f \"C:\\Users\\jschulz\\Dropbox\\Programme\\cmder\\vendor\\jasc\\nbflatten.jq\" --arg show_output 0"
cachetextconv = false
If I have notebooks in a repo which I want cleaned up before committing and/or diffing, I add a .gitattribute file with the following content:
*.ipynb filter=stripoutput
*.ipynb diff=ipynb
Please note that both together mean that the ipynb git diff engine never sees the output in a notebook (as the filter is run before the diff), so most of the above nbflatten.jq file is useless in that case (and even without the filter it would still not show up until you change “show_output 0” to “show_output 1”) . But you can use it via an alias (in <cmder>\config\aliases) ala
nbflat=jq -r -f "C:\Users\jschulz\Dropbox\Programme\cmder\vendor\jasc\nbflatten.jq" --arg show_output 1 $*
and then use it like nbflat whatever.ipynb | less to get a text representation.
nbconvert
I installed nbconvert into the main conda env: deactivate & conda install nbconvert
For pdf output, I installed miktex and pandoc:
- miktex: latex environment. Installed via the portable installer and added to the path (via
setx path c:\path\to\MIKTEX\miktex\bin;%path% in a cmd window, not cmder -> that way you have latex available in all programs and not only in a cmder window).
- pandoc: converter between text formats (e.g. markdown to word or pdf). Also added to the path like miktex.
It has to go to the main path (not setup via cmder), as the way I startup a notebook server does not get the path additions from cmder…
Other stuff
- everything: search for filenames (not content). Installed as a service and then put
es.exe in a dir in %PATH% (e.g. <cmder>\bin). es whatever.py will now show all files with that name.
- launchy: search and startup commands fast. Faster than
Start-><search box>-><Enter>… I used that much more when I had WinXP installed. Nowadays, I have most programs added as a shortcut to the quickstart area.
- Chrome with ublock (ad blocking) and The Great Suspender (suspend tabs which you haven’t touched in days so that they don’t waste resources).
- sysinternals:
procexplorer (graphical process explorer, replacement for the task manager). Setup to start as admin during windows startup. I also use autostarts from time to time to clean up the autostart entries.
- Keepass 2: holds all my passwords, integrated with chrome via chromeIPass. The keepass file is synced via dropbox to my mobile (+ a keyfile which is manually transfered…).
So, you can make yourself at home on windows as a (python) developer… Unfortunately, it seems that there are not a lot of people who do dev work on windows (based on the many projects which fail on windows when I check them out). If you want to make your project windows friendly: add Appveyor to your CI tests… :-)
Anyway: anything I missed to make my life on windows any easier?
Jan 24, 2016
Conda recipes can contain patches which are applied on top of the source for the package. When updating the package to a new upstream version, these patches need to be checked if the still apply (or are still needed).
This is the way I do it currently (be aware that I work on windows, so you might need to change some slashes…)…
Preparation
# makes the "patch" command available...
set "PATH=%path%;C:\Program Files\Git\usr\bin\"
# Update the latest source for matplotlib...
cd matplotlib
git fetch
git checkout origin/master
# conda package recipe for matplotlib is in ci\conda_recipe
Apply a patch
patch -i ci\conda_recipe\osx-tk.patch
The next step depends whether the patch applied cleanly or not. There are three possible outcomes:
- The patch applied cleanly (e.g. no error message): nothing further to do, on to the next patch…
- The patch is fuzzy (
Hunk #1 succeeded at 1659 with fuzz 1 (offset 325 lines).) -> the patch only needs to be refreshed
- The patch (or one of the hunks) didn’t apply (
1 out of 1 hunk FAILED -- saving rejects to file matplotlibrc.template.rej) -> the patch needs to be redone and afterwards the patch needs to be refreshed
For redoing the patch, look into the <patch>.rej file and apply similar changes to the source. Or check whether this patch is needed anymore…
For refreshing the patch, make sure that only the changes for the patch are currently included in you checked out copy (e.g. make sure that refreshed patches are git added before the next command…).
Then run the following command:
git diff --no-prefix > ci\conda_recipe\osx-tk.patch
[I actually used a different filename to pipe the patch to and then compared the output before overwriting the old patch…]
Nov 14, 2015
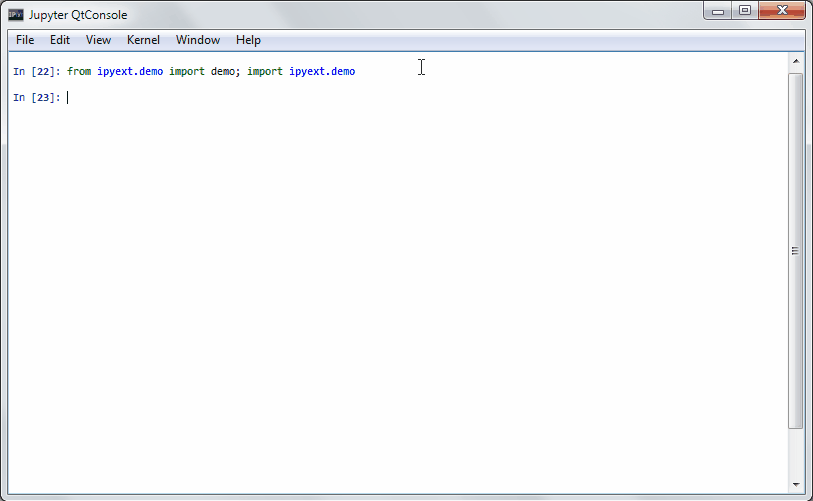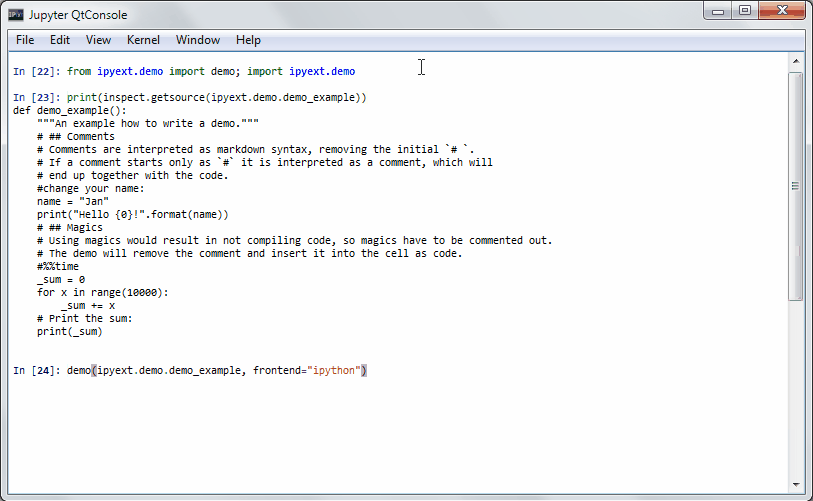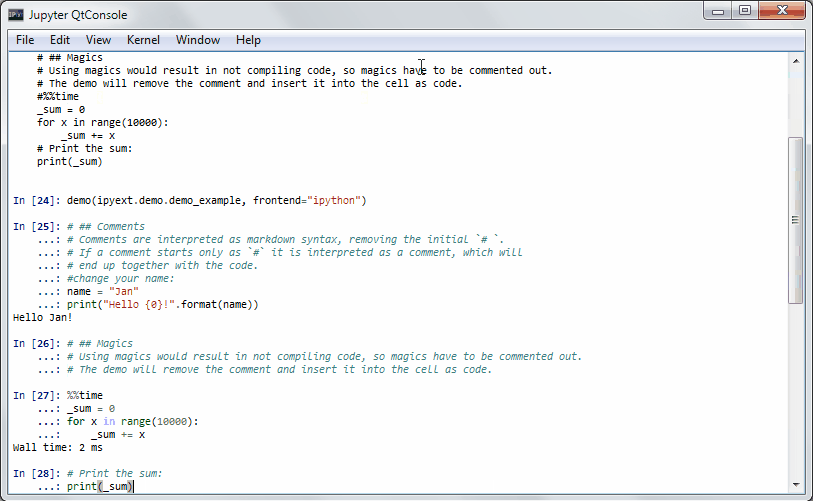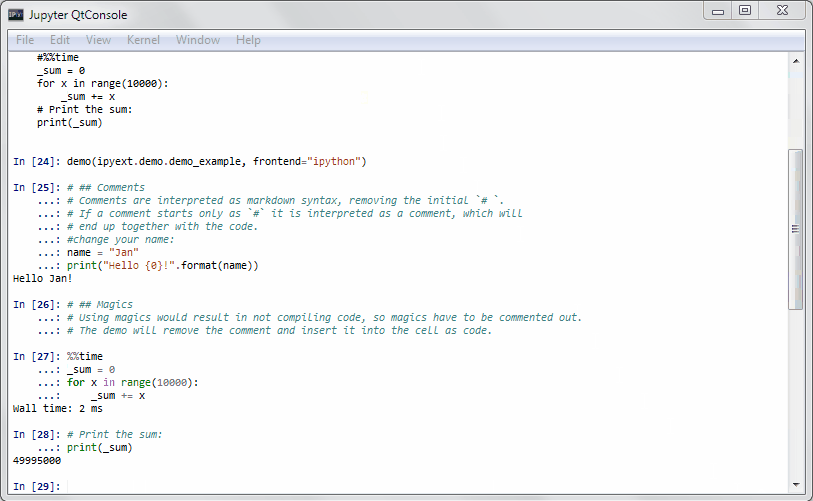
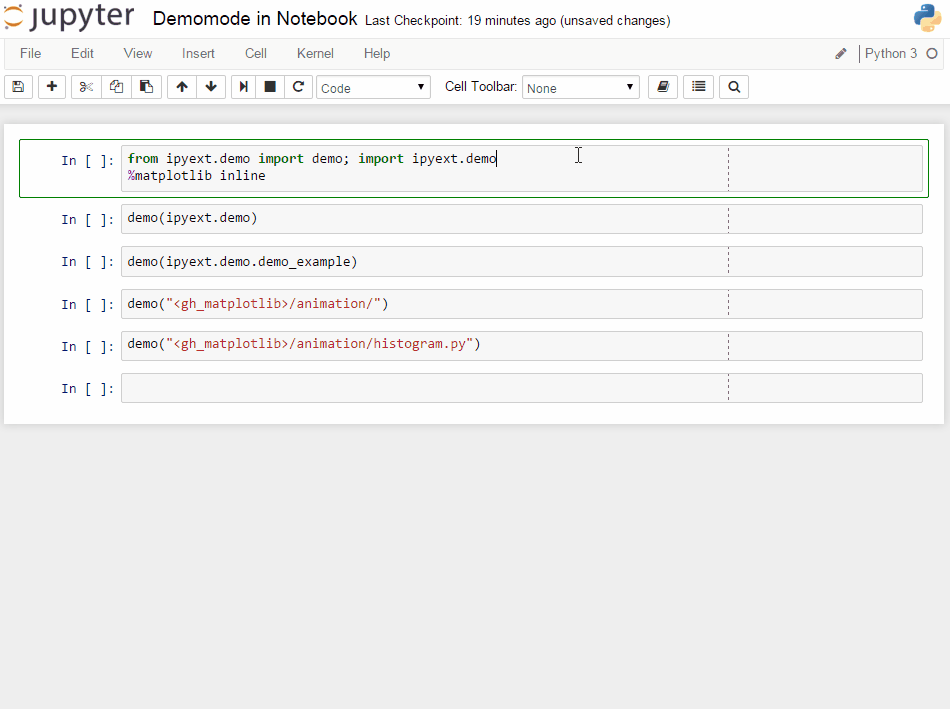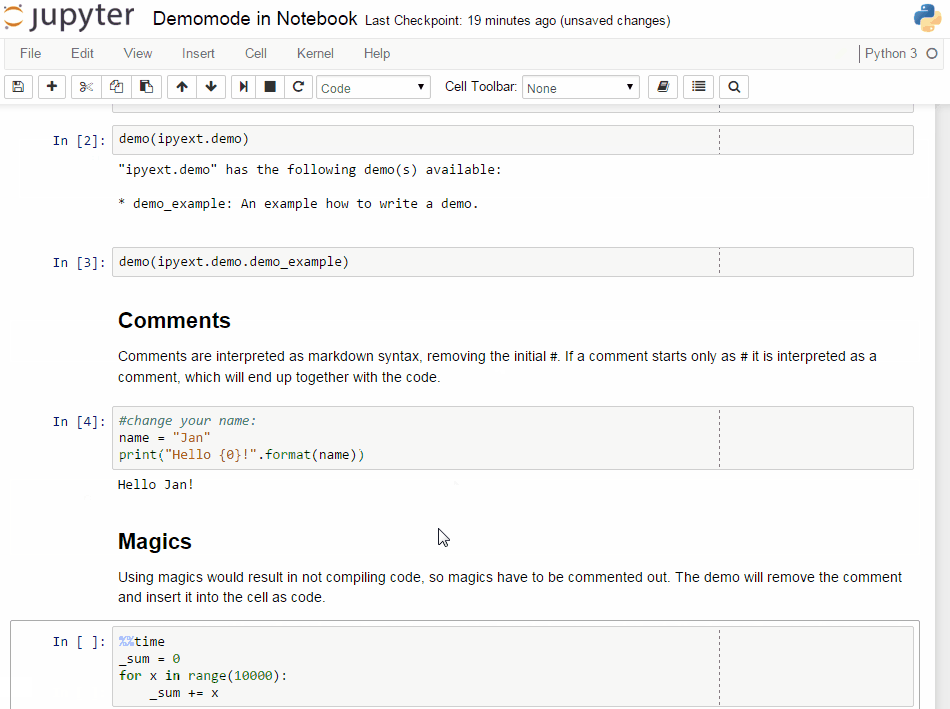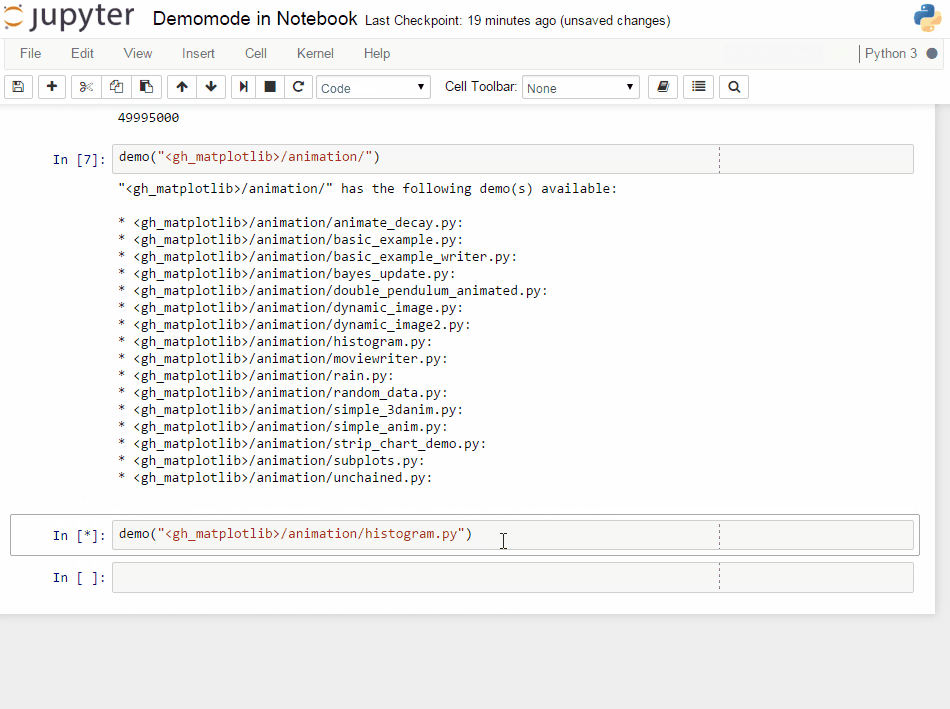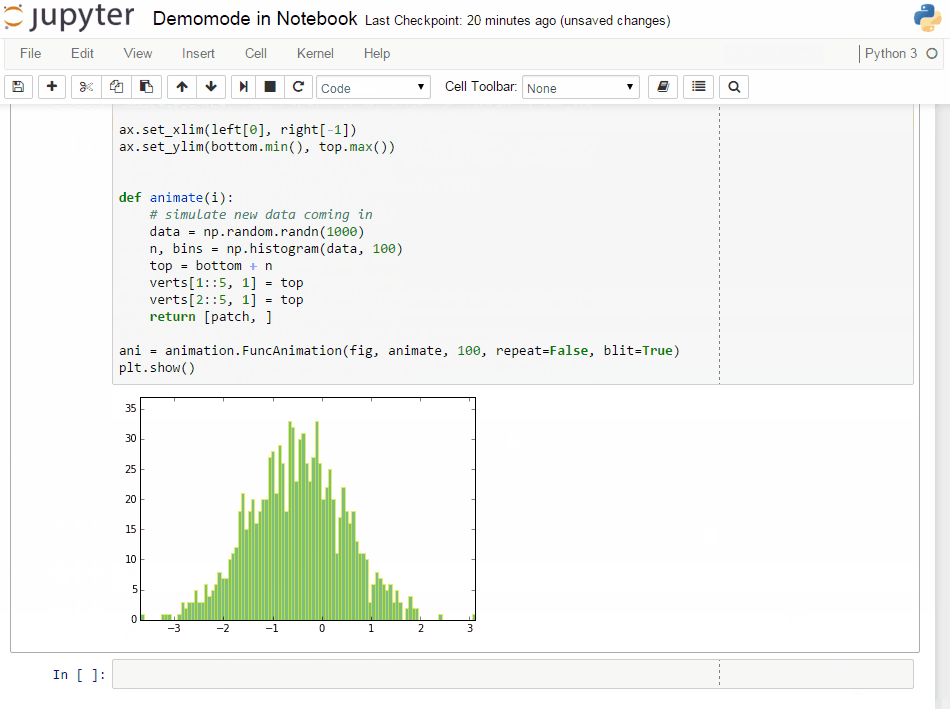
R has a demo mode, which lets you execute some demo of a function or a package. See e.g. demo(lm.glm) for such a thing.
An PR in IPython-extensions lets you do much the same:
It will get some demo code (which can be a function in a package or the matplotlib examples on github) and lets you execute that code by yourself. Specially formatted comments in the function will get turned into formatted text, if the frontend suppports it. It works in the notebook by adding new cells with the demo content or in the qtconsole/ipython directly by presetting it as new input (simple press enter) until the demo is over.
Writing a demo
Writing a demo is simple writing a function in a module. Markdown formatting in comments is possible and works in the notebook. In the qtconsole/IPython, they are simple comments.
This is the demo example:
[...]
def demo_example():
"""An example how to write a demo."""
# ## Comments
# Comments are interpreted as markdown syntax, removing the
# initial `# `. If a comment starts only with `#`, it is interpreted
# as a code comment, which will end up together with the code.
#change your name:
name = "Jan"
print("Hello {0}!".format(name))
# ## Magics
# Using magics would result in not compiling code, so magics
# have to be commented out. The demo will remove the comment
# and insert it into the cell as code.
#%%time
_sum = 0
for x in range(10000):
_sum += x
# Print the sum:
print(_sum)
# This lets the `demo(ipyext.demo)` find only the `demo_example`.
# Only modues with that variable will display an overview of
# the available demos.
__demos__ = [demo_example]
Demo of demo mode :-)
Here are some videos of it in action:
IPython qtconsole
Jupyter Notebook (with IPython kernel)
If you have any comments, or know of examples for a package (needs to be plain python files available on github -> like for
matplotlib), please leave it below or in the
PR. Thanks!
Oct 22, 2015
Recently I found the conda-forge project on github which makes it easy to auto build and upload your python project as a (native) conda package. Conda-forge introduces the concept of a “smithy” (a repository on github) which builds the conda packages for the main repository. A smithy connects to three different CI services to get builds for all three major platforms: Travis for Mac OS X, CircleCI for Linux and AppVeyor for Windows.
But not everyone is using conda environments so I thought it would be nice to to also build wheels with it. Turns out this is actually possible, you “only” need to hack the conda build scripts to also build a wheel and upload that to PyPI.
For those who just want the code: you can find it in the JanSchulz/package-builder repo on github. The smithy currently builds the conda packages and wheels for pypandoc. PRs welcome :-)
These were the broad steps to setup the smithy:
- Setup a smithy repository: I copied the conda-forge/staged-recipes, which is setup to build multiple recipes, but required a little more “handholding” until I got it to run (the “one main repo, one smithy repo” case has helper scripts in conda-forge/conda-smithy, which will make the setup of the smithy a lot easier. So try that first if that fits your case…):
- Add your own conda recipe: it’s usually three easy files in a subdir: one metadata file (
meta.yaml) and one script for each windows (bld.bat) and unix-like platforms (build.sh). Take a look at some examples…
- Connect the various CI services to your github repo.
- Get your anaconda access TOKEN via
anaconda auth --create --name WhateverName --scopes "repos conda api" (I used a different token for each CI service). The examples in the conda-forge/staged-recipes files didn’t quite work, as I needed to add api access…
- Add your anaconda access TOKEN to each CI service so that it is available in your build environment.
- Hack your conda recipe to also build a wheel and upload that to PyPI. This is a bit more involved, as conda builds happen in a temporary environment and have their environment variables cleaned up. So:
- Install twine in the environment, by adding
pip install twine to the various CI setup scripts (unfortunately it’s not packaged for conda, so you can’t simple install it via meta.yaml).
- Add your PyPI username and password as a environment variable to each CI service.
- Let
conda build know that you want to have these two variables available during the conda build by adding them to the build -> script_env section of your meta.yaml.
- Add a line to your build scripts to build a wheel (
python setup.py bdist_wheel).
- Generate a
pypirc file so that the PyPI upload can happen. This is a bit tricky, as the build process has no access to the recipe directory and therefore you have to generate this file on the fly during build. On unix-like it’s a cat << EOF > pypirc\n...\nEOF, but on windows you have to use either a lot of echo ... >>pypirc or a trick with parenthesis: ( echo ...; echo ... ...) > "pypirc". It seems that twine doesn’t work without such a file :-(.
- Use twine to upload the package: this currently means that you have to add a username and password (using the added environment variables) to the commandline, so make sure that this line isn’t echo’ed to the CI log: use
@twine ... in bld.bat and set +x; twine ...; set -x in build.sh.
- I also added a test to
build.sh to only build wheels on darwin, as it seems that PyPI does not accept linux wheels…
- Fix all the errors you introduced and repush the repo… this step took a bit… :-/
Now making a release works like this:
- Release the package (in my case pypandoc) as usual.
- Build and upload the
sdist to PyPI.
- Update the conda recipe for the new version.
- Push the smithy repo with the recipe and let the CI services build the conda packages and the wheels.
The CI scripts will only build packages for which there are no current conda packages yet. If you need to redo the build because of some problems, you need to delete the conda packages for the affected builds or bump the package versions (you can set build versions for the conda packages without bumping the version of the upstream project).
If you have any feedback, please leave it in the comments (or as an issue in one of the above repos… :-) ).
The next step will be adding builds for R packages…
Sep 18, 2015
I recently had to explore a JSON API and came up with the following twothree functions to make working with the returned JSON/dict easier:
[Update 2015-11-10: you might like dripper, which does much of this code snippet…]
[Update 2015-09-26: updates to code and new convert_to_dataframe_input function: see here for a post about it]
_null = object()
def get_from_structure(data, name, default=_null):
"""Return the element with the given name.
`data` is a structure containing lists, dicts or scalar values.
A name is a '.' separated string which specifies the path in the data.
E.g. '0.name.first' would return `data[0]["name"]["first"]`.
If such a path does not exist and no default is given, a
KeyError is raised. Otherwise, the default is returned.
"""
names = name.split(".")
for n in names:
try:
i = int(n)
data = data[i]
except:
data = data.get(n, _null)
if data is _null:
if default is _null:
raise KeyError("Key not found: %s (%s)" % (n, name))
else:
return default
return data
def find_in_structure(data, value):
"""Find the value in the data and returns a name for that element.
`value` is either found by "==" (elements are equal) or "in" (part of
a string or other element in an iterable other than list).
The name is a '.' separated path (string) suitable for `get_from_dict`.
Raises a ValueError if the value is not found in data.
"""
_stack = []
def _find(data, stack):
if data is None:
return False
if isinstance(data, list):
for i, val in enumerate(data):
stack.append(str(i))
if _find(val, stack):
return True
else:
stack.pop()
elif isinstance(data, dict):
for key, val in data.items():
stack.append(key)
if _find(val, stack):
return True
else:
stack.pop()
elif data == value or value in data:
return True
return False
if _find(data, _stack):
return ".".join(_stack)
else:
raise ValueError("Not found in data: %s" % (text,))
def convert_to_dataframe_input(data, converter_dict):
"""Convert the input data to a form suiteable for pandas.Dataframe
Each element in data will be converted to a dict of key: values by using
the functions in converter_dict. If feed to a pandas.DataFrame, keys
in converter_dict will become the column names.
If an element in converter_dict is not callable, it will be used
as an name for `get_from_dict`. If the function raises an Exception,
NA will be filled in.
If data is a dict, the key will be used for a `_index` column,
otherwise a running index is used.
This function does not do any type conversations.
"""
from functools import partial
NA = float('nan')
converted = []
assert '_index' not in converter_dict, "'_index' is not allowed as a key in converter_dict"
temp = {}
for k, val in converter_dict.items():
if not callable(val):
temp[k] = partial(get_from_structure, name=val)
else:
temp[k] = val
converter_dict = temp
if isinstance(data, dict):
gen = data.items()
else:
gen = enumerate(data)
for index, item in gen:
d = {"_index": index}
for name, func in converter_dict.items():
try:
d[name] = func(item)
except:
d[name] = NA
converted.append(d)
return converted
Examples:
data = {"ID1":{"result":{"name":"Jan Schulz"}},
"ID2":{"result": {"name":"Another name", "bday":"1.1.2000"}}}
print(find_in_structure(data, "Schulz"))
## ID1.result.name
print(get_from_structure(data, find_in_structure(data, "Schulz")))
## Jan Schulz
And the DataFrame conversion
converter_dict = dict(
names = "result.name",
bday = "result.bday"
)
import pandas as pd
print(pd.DataFrame(convert_to_dataframe_input(data, converter_dict)))
## _index bday names
## 0 ID1 NaN Jan Schulz
## 1 ID2 1.1.2000 Another name
Someone might find this useful (and at least I can find it again :-) )

